AI protein prediction tool AlphaFold3 now available as open source
DeepMind's AlphaFold3 is now open source. Researchers can use the AI protein structure prediction tool for non-commercial purposes.
AI protein prediction tool AlphaFold3 now available as open source
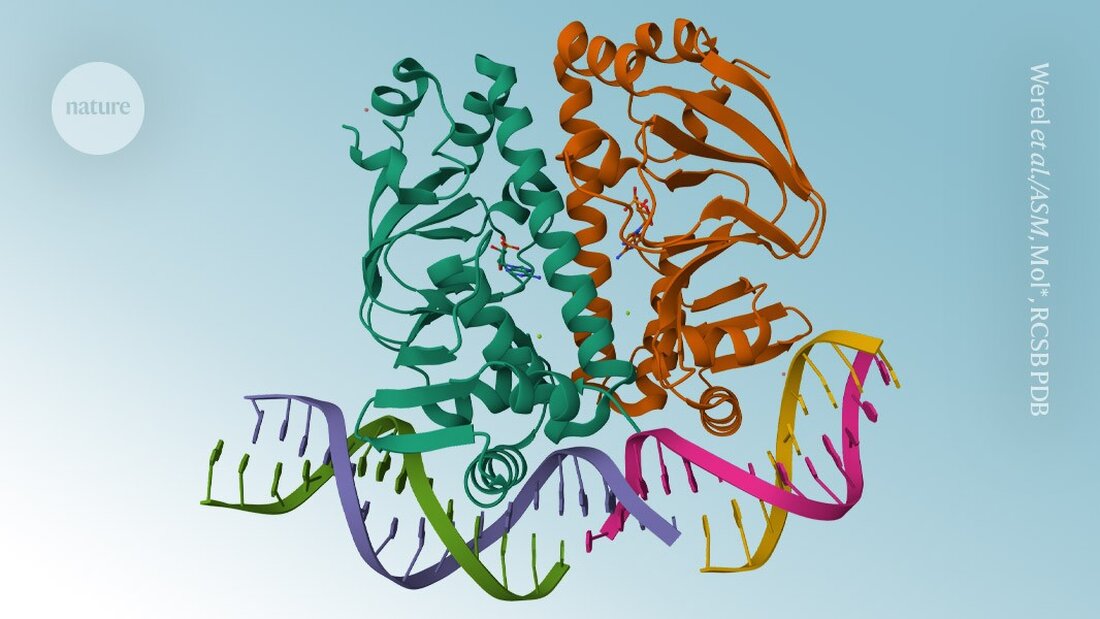
AlphaFold3 is finally available. Six months after Google DeepMind controversial the code one Papers on the protein structure prediction model scientists can now download the software code and use the artificial intelligence tool for non-commercial applications, the London-based company announced on November 11.
“We’re very excited to see what people do with it,” says John Jumper, who leads the AlphaFold team at DeepMind and joined CEO Demis Hassabis last month won part of the 2024 Nobel Prize in Chemistry for their work on the AI tool.
Unlike its predecessors, AlphaFold3 able to model proteins in combination with other molecules. Instead of releasing the underlying code - as is the case with AlphaFold2 was the case – DeepMind provided access via a web server that limited the number and type of predictions scientists could make.
Critically, the AlphaFold3 server did not allow scientists to predict how proteins would react in the presence of potential drugs. But now DeepMind's decision to release the code means academic scientists can predict such interactions by running the model themselves.
The company initially stated that making AlphaFold3 available only through a web server struck the right balance between providing access for research and protecting commercial ambitions. Isomorphic Labs, a spin-off of DeepMind in London, is applying AlphaFold3 in drug discovery.
However pulled the release of AlphaFold3 without its code or model weights — Parameters obtained by training the software on protein structures and other data — drew criticism from scientists who said the move undermined reproducibility. DeepMind quickly drew conclusions and said that an open source version of the tool would be made available within six months.
Anyone can now download the AlphaFold3 software code and use it non-commercially. However, currently only scientists with academic affiliations have access to the training weights upon request.
Accessible versions
DeepMind has competition: In recent months, several companies have Open source tools for protein structure prediction based on AlphaFold3 presented, which rely on specifications described in the original paper, known as pseudocode.
Two Chinese companies — tech giant Baidu and TikTok developer ByteDance — have released their own AlphaFold3-inspired models, as has a startup in San Francisco, California called Chai Discovery.
A key drawback of these models is that none of them, like AlphaFold3, are licensed for commercial applications such as drug discovery, says Mohammed AlQuraishi, a computational biologist at Columbia University in New York City. However, Chai Discovery's model, Chai-1, can be used for such work via a web server, explains Jack Dent, the company's co-founder.
Another company, Ligo Biosciences of San Francisco, has released a restriction-free version of AlphaFold3. However, this does not yet have the full range of functions, including the ability to model drugs and molecules other than proteins.
Other teams are working on versions of AlphaFold3 that are available without such restrictions: AlQuraishi hopes to offer a fully open-source model called OpenFold3 later this year. This would allow pharmaceutical companies to retrain their own versions of the model using proprietary data, such as the structures of proteins bound to different drugs, potentially improving performance.
Openness counts
The past year has seen a rush of new biological AI models from companies with different approaches to openness. Anthony Grid, a computational biologist at the University of Wisconsin-Madison, has no problem with commercial companies entering his field—as long as they follow the same rules as other scientists when sharing their work in journals and preprint servers.
If DeepMind makes claims about AlphaFold3 in a scientific publication, "I would expect them to also share information about how the predictions were made and provide the AI models and code in a way that we can test them," grid adds. “My group will not use tools that we cannot verify.”
The fact that several replications of AlphaFold3 have already emerged shows that the model was reproducible, even without open source code, says Pushmeet Kohli, the head of AI for Science at DeepMind. He adds that he would like to see more discussion in the future about publishing standards in a field increasingly populated by academic and corporate researchers.
The open source nature of AlphaFold2 led to a surge of innovation from other scientists. For example, the winners of a recent protein modeling competition used the AI tool to to design new proteins that can bind to a cancer target. Jumper's favorite AlphaFold2 hack comes from a team that used the tool to to identify an important protein that helps sperm attach to eggs.
Jumper can't wait to see what surprises emerge after the release of AlphaFold3 - even if they aren't always successful. “People will use it in strange ways,” he predicts. “Sometimes it will fail and sometimes it will succeed.”

 Suche
Suche
 Mein Konto
Mein Konto
